Publications
Publications from our group.
Highlighted
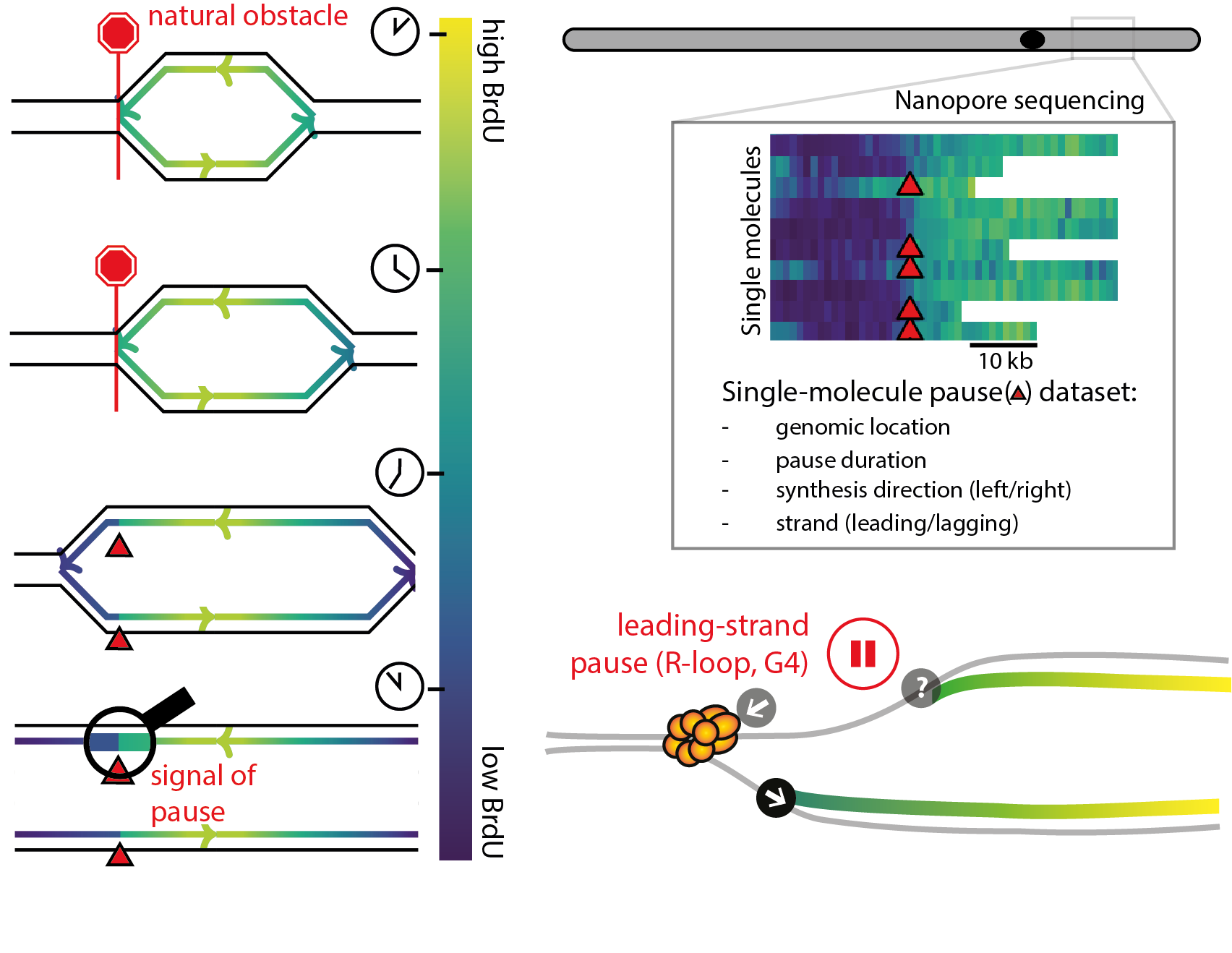
Single-molecule landscape of DNA replication pausing
bioRxiv
·
14 Aug 2025
·
doi:10.1101/2025.08.14.670160
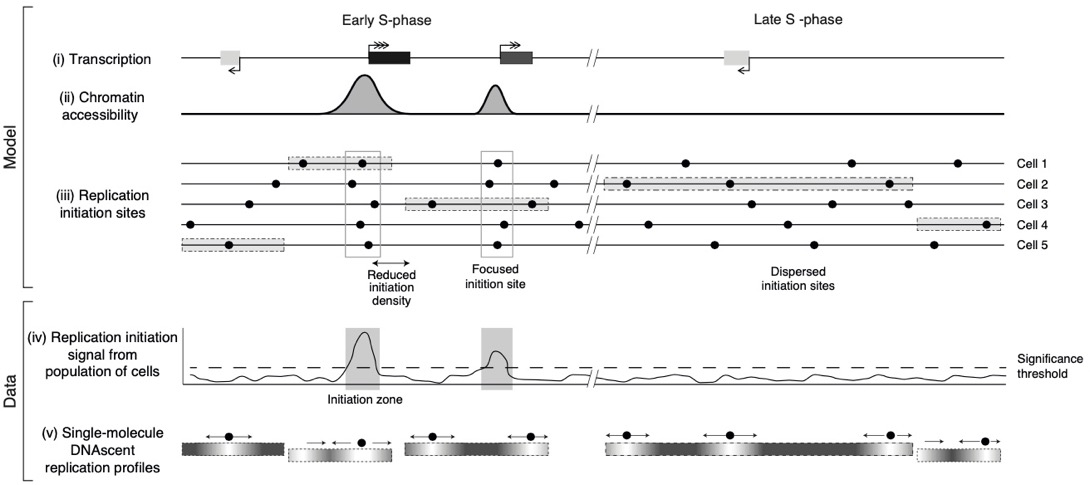
Most human DNA replication initiation is dispersed throughout the genome with only a minority within previously identified initiation zones
Genome Biology
·
09 May 2025
·
doi:10.1186/s13059-025-03591-w
Discovering where DNA replication initiates in human cells is crucial for understanding how cells copy their genetic information. Our study explores where and how often DNA replication begins across the human genome. To achieve this, we employed a technique using in vivo BrdU incorporation followed by detection by single-molecule nanopore sequencing. We tested various BrdU concentrations on cultured human cells, sequenced the resulting DNA, and developed a computational method to identify replication initiation events from the sequencing data. We discovered that a large proportion of DNA replication begins outside known initiation zones, demonstrating a dispersed initiation pattern across the genome. We concluded that while a subset of initiation events occurs in focused zones and is influenced by factors such as transcription, the majority are dispersed and not associated with specific sequence features or epigenetic marks. This understanding refines the current model of how DNA replication is orchestrated within human cells and could inform studies on genome stability and the onset of diseases like cancer.

Design, construction, and functional characterization of a tRNA neochromosome in yeast
Cell
·
01 Nov 2023
·
doi:10.1016/j.cell.2023.10.015
All
2025

Single-molecule landscape of DNA replication pausing
bioRxiv
·
14 Aug 2025
·
doi:10.1101/2025.08.14.670160
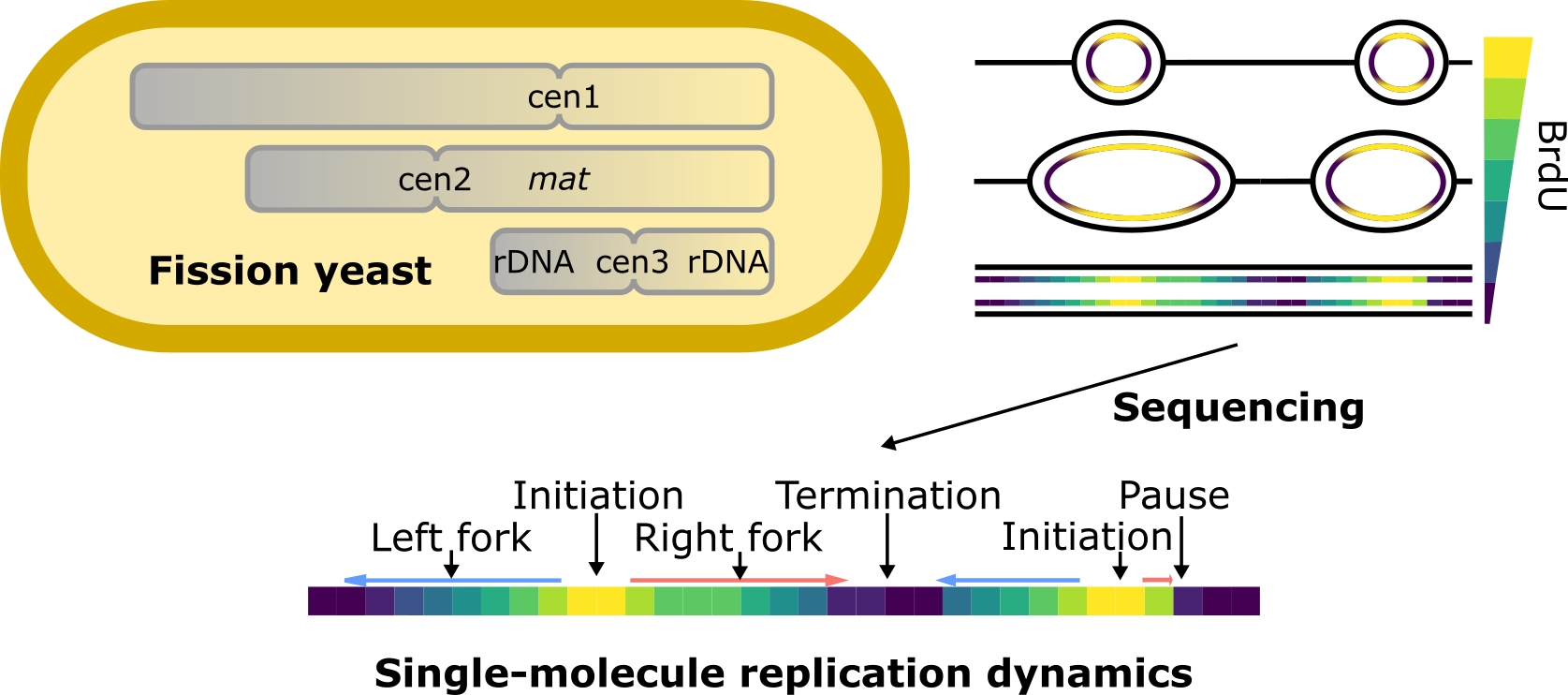
Single-molecule sequencing maps replication dynamics across the fission yeast genome, including centromeres
bioRxiv
·
18 Jul 2025
·
doi:10.1101/2025.07.16.665067

Most human DNA replication initiation is dispersed throughout the genome with only a minority within previously identified initiation zones
Genome Biology
·
09 May 2025
·
doi:10.1186/s13059-025-03591-w
Discovering where DNA replication initiates in human cells is crucial for understanding how cells copy their genetic information. Our study explores where and how often DNA replication begins across the human genome. To achieve this, we employed a technique using in vivo BrdU incorporation followed by detection by single-molecule nanopore sequencing. We tested various BrdU concentrations on cultured human cells, sequenced the resulting DNA, and developed a computational method to identify replication initiation events from the sequencing data. We discovered that a large proportion of DNA replication begins outside known initiation zones, demonstrating a dispersed initiation pattern across the genome. We concluded that while a subset of initiation events occurs in focused zones and is influenced by factors such as transcription, the majority are dispersed and not associated with specific sequence features or epigenetic marks. This understanding refines the current model of how DNA replication is orchestrated within human cells and could inform studies on genome stability and the onset of diseases like cancer.
2024
Most human DNA replication initiation is dispersed throughout the genome with only a minority within previously identified initiation zones
bioRxiv
·
01 May 2024
·
doi:10.1101/2024.04.28.591325

Telomere‐to‐telomere Schizosaccharomyces japonicus genome assembly reveals hitherto unknown genome features
Yeast
·
01 Mar 2024
·
doi:10.1002/yea.3912
2023

Schizosaccharomyces versatilis represents a distinct evolutionary lineage of fission yeast
Yeast
·
26 Dec 2023
·
doi:10.1002/yea.3919

Design, construction, and functional characterization of a tRNA neochromosome in yeast
Cell
·
01 Nov 2023
·
doi:10.1016/j.cell.2023.10.015
2022

Systematic Analysis of Copy Number Variations in the Pathogenic Yeast Candida parapsilosis Identifies a Gene Amplification in
RTA3
That is Associated with Drug Resistance
mBio
·
26 Oct 2022
·
doi:10.1128/mbio.01777-22
2021
Resistance to miltefosine results from amplification of the RTA3 floppase or inactivation of flippases in Candida parapsilosis
Cold Spring Harbor Laboratory
·
17 Dec 2021
·
doi:10.1101/2021.12.16.473093
Effectiveness of glass beads for plating cell cultures
Physical Review E
·
17 May 2021
·
doi:10.1103/PhysRevE.103.052410

Tos4 mediates gene expression homeostasis through interaction with HDAC complexes independently of H3K56 acetylation
Journal of Biological Chemistry
·
01 Jan 2021
·
doi:10.1016/j.jbc.2021.100533
2020

Sir2 mitigates an intrinsic imbalance in origin licensing efficiency between early- and late-replicating euchromatin
Proceedings of the National Academy of Sciences
·
08 Jun 2020
·
doi:10.1073/pnas.2004664117
This work is H1 recommended.
The Beacon Calculus: A formal method for the flexible and concise modelling of biological systems
PLOS Computational Biology
·
09 Mar 2020
·
doi:10.1371/journal.pcbi.1007651
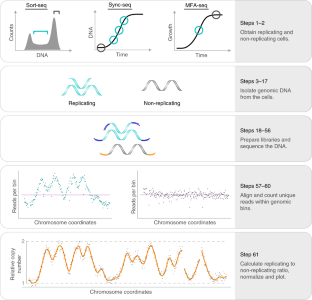
DNA copy-number measurement of genome replication dynamics by high-throughput sequencing: the sort-seq, sync-seq and MFA-seq family
Nature Protocols
·
12 Feb 2020
·
doi:10.1038/s41596-019-0287-7
During S phase, the relative DNA copy number increases for each region as it is replicated. Therefore, measuring DNA copy number can be used as a proxy for replication time. Here we present a step-by-step protocol to determine genome replication dynamics using high-throughput DNA sequencing to measure DNA copy-number. Cell populations can be obtained in three variants of the method sort-seq, sync-seq and MFA-seq.
We have shared our computational pipeline and example sequence data - see the resources page.
We have shared our computational pipeline and example sequence data - see the resources page.
2019
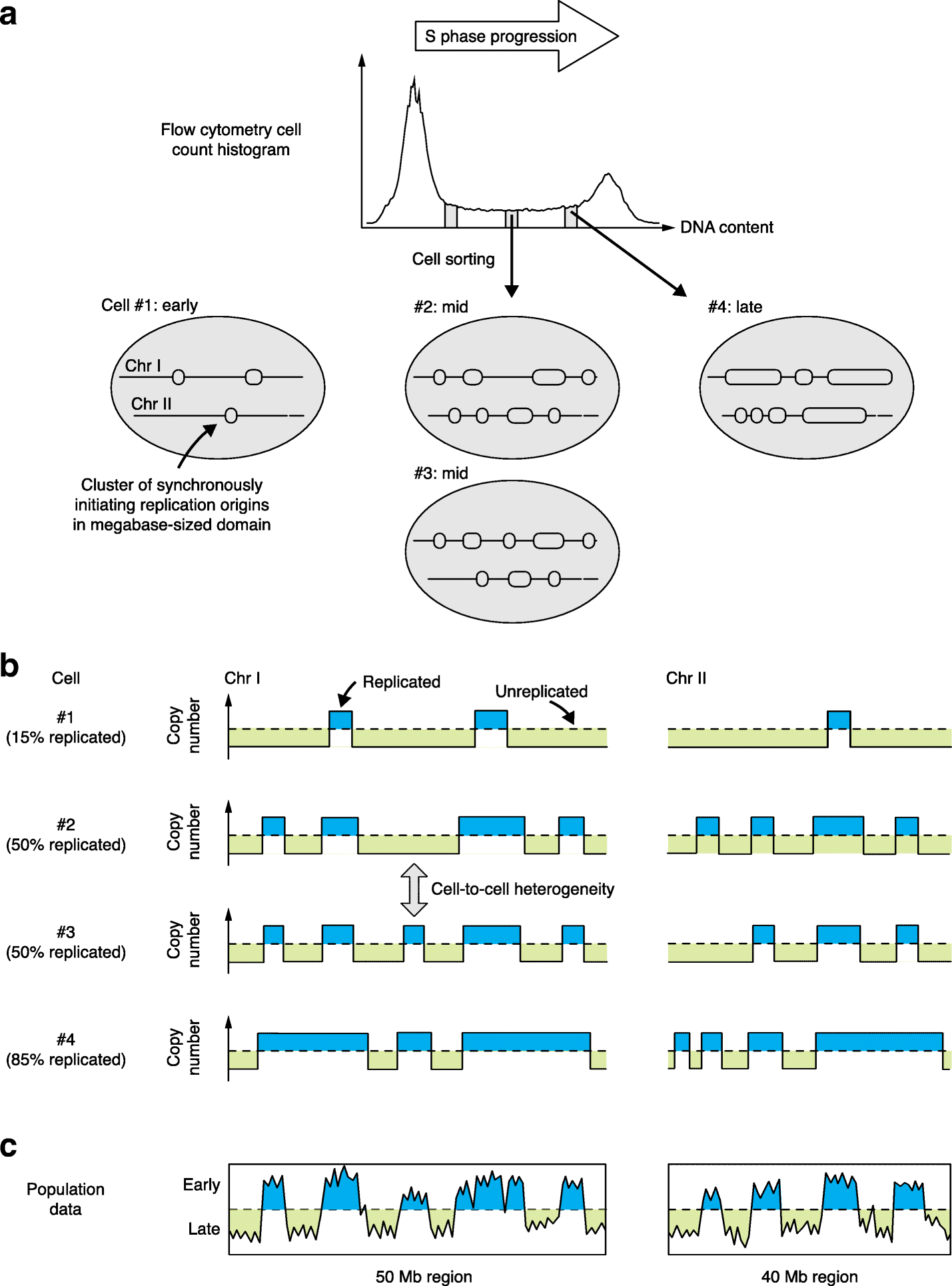
Genome-wide analysis of DNA replication timing in single cells: Yes! We’re all individuals
Genome Biology
·
30 May 2019
·
doi:10.1186/s13059-019-1719-y
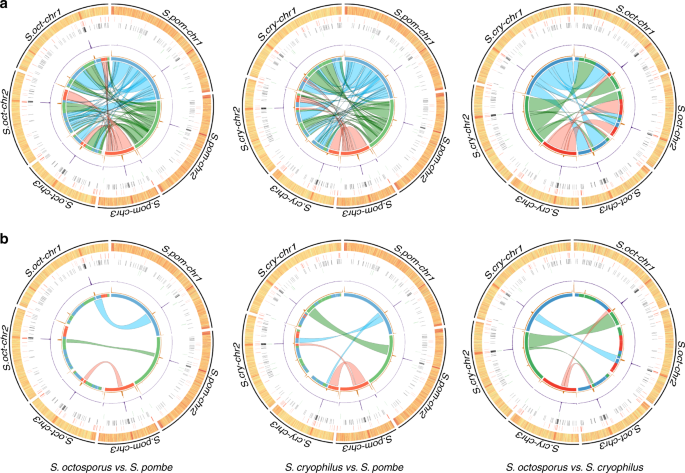
Interspecies conservation of organisation and function between nonhomologous regional centromeres
Nature Communications
·
28 May 2019
·
doi:10.1038/s41467-019-09824-4
This work is H1 recommended.
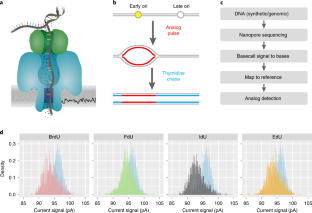
Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads
Nature Methods
·
22 Apr 2019
·
doi:10.1038/s41592-019-0394-y
DNA replication initiation is stochastic and obscured in population datasets. In this paper we introduce D-NAscent, a method to detect base analogs in ultra-long nanopore sequencing reads. This allowed us to identify replication origins and replication fork movement genome-wide on sequence reads of 20–160 kilobases. Our study marks the first, whole genome characterisation of DNA replication dynamics on single-molecules.
We have shared both the D-NAscent software and underlying sequence data - see the resources page.
This work is H1 recommended.
We have shared both the D-NAscent software and underlying sequence data - see the resources page.
This work is H1 recommended.
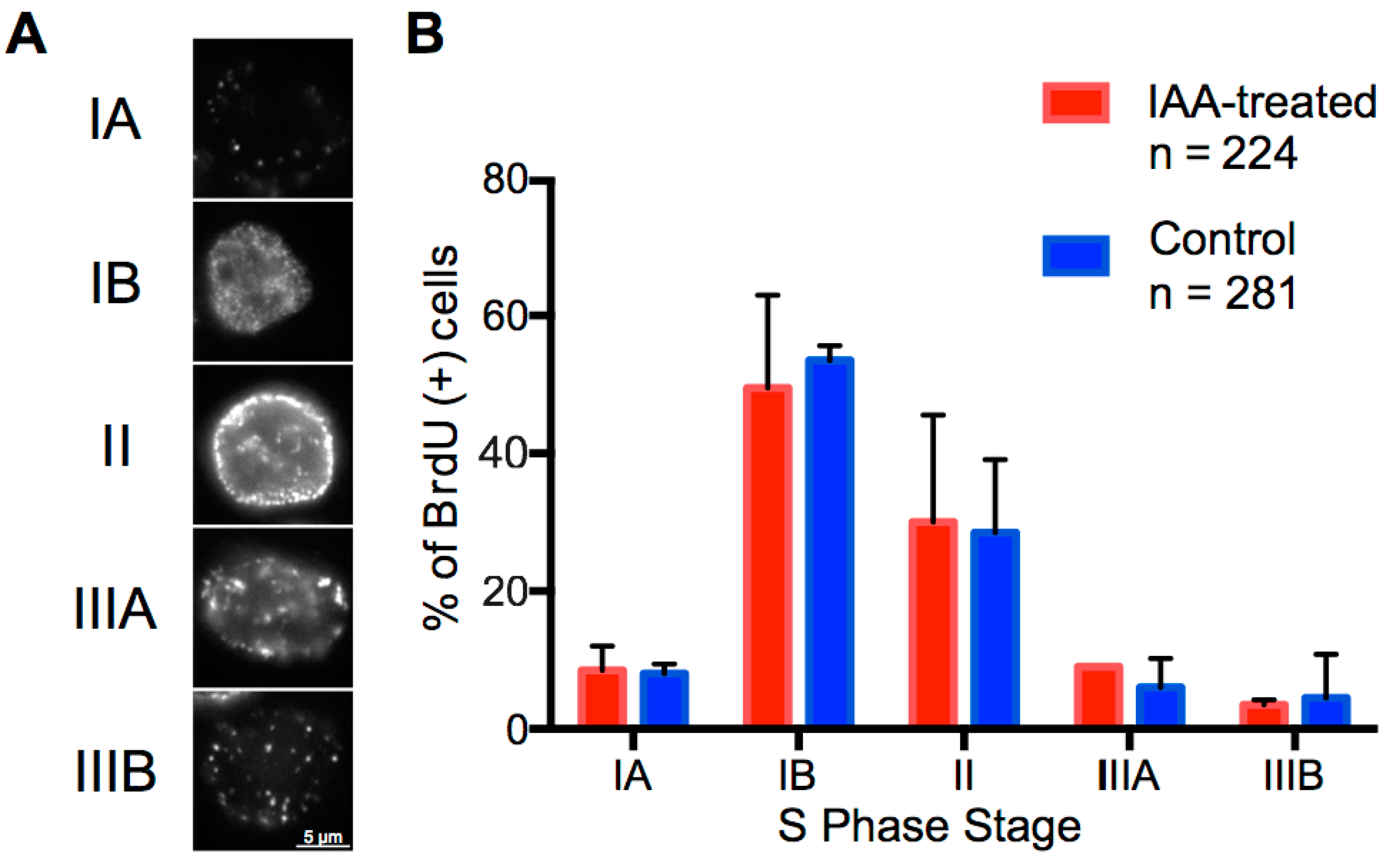
Cohesin-Mediated Genome Architecture Does Not Define DNA Replication Timing Domains
Genes
·
04 Mar 2019
·
doi:10.3390/genes10030196
In this study we address the long-standing question - do topologically associating domains (TADs) act as a causative organizing structure for replication timing domains? Using rapid and complete cohesin degradation we observe no perturbations to replication timing. Our results suggest that cohesin-mediated genome architecture is not required for replication timing patterns.
Bayesian inference of origin firing time distributions, origin interference and licencing probabilities from Next Generation Sequencing data
Nucleic Acids Research
·
14 Feb 2019
·
doi:10.1093/nar/gkz094
2018
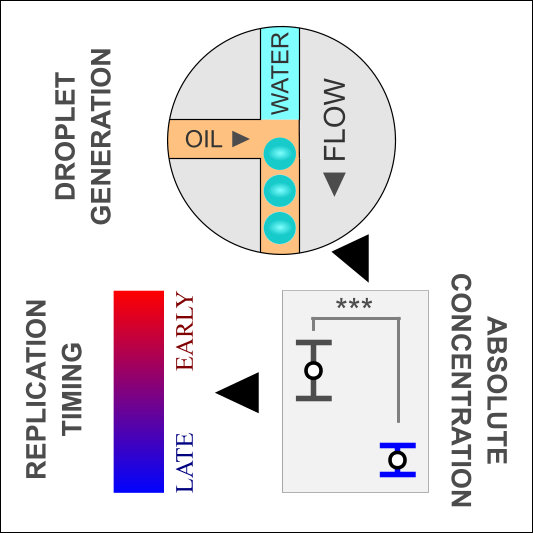
Rapid high-resolution measurement of DNA replication timing by droplet digital PCR
Nucleic Acids Research
·
09 Jul 2018
·
doi:10.1093/nar/gky590
In this paper we describe the use of digital PCR to precisely and rapidly measure DNA copy number as a proxy for replication time. As a proof-of-principle we demonstrate the method’s utility for the study of DNA replication time in both yeast and human cells. We show the power of the method by undertaking a targeted screen of kinetochore mutants to discover those that affect centromere replication time. As part of this work we generated whole genome DNA replication timing data (sort-seq) for cultured human cells (HeLa).
These data can be visualised via the link to our UCSC genome browser hub on the resources page.
These data can be visualised via the link to our UCSC genome browser hub on the resources page.

Evolution of Genome Architecture in Archaea: Spontaneous Generation of a New Chromosome in Haloferax volcanii
Molecular Biology and Evolution
·
16 Apr 2018
·
doi:10.1093/molbev/msy075
Investigating the role of Rts1 in DNA replication initiation
Wellcome Open Research
·
06 Mar 2018
·
doi:10.12688/wellcomeopenres.13884.1

Rif1 acts through Protein Phosphatase 1 but independent of replication timing to suppress telomere extension in budding yeast
Nucleic Acids Research
·
26 Feb 2018
·
doi:10.1093/nar/gky132
The effectiveness of glass beads for plating cell cultures
Cold Spring Harbor Laboratory
·
01 Jan 2018
·
doi:10.1101/241752
2017
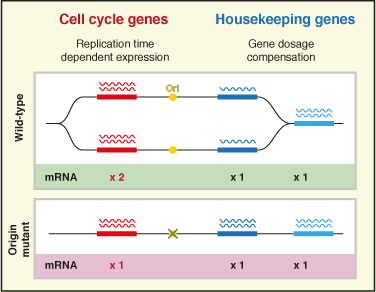
DNA replication timing influences gene expression level
Journal of Cell Biology
·
24 May 2017
·
doi:10.1083/jcb.201701061
Chromosomes replicate in a characteristic and reproducible temporal order. However, it has been unknown whether there is a physiological requirement for the regulation of replication timing. In this paper, we report our discovery that early DNA replication is required for the highest expression levels of certain genes.
Highlighted in J. Cell Biol. by María Gómez, selected for the Nuclear biology special collection by Ana Pombo and selected for the DNA replication and genome instability special collection by Agata Smogorzewska.
Selected as “of outstanding interest” by Mirit Aladjem and colleagues in their Current Opinion in Cell Biology review.
Highlighted in J. Cell Biol. by María Gómez, selected for the Nuclear biology special collection by Ana Pombo and selected for the DNA replication and genome instability special collection by Agata Smogorzewska.
Selected as “of outstanding interest” by Mirit Aladjem and colleagues in their Current Opinion in Cell Biology review.
Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome
Science
·
10 Mar 2017
·
doi:10.1126/science.aaf4791
2016
Global and Local Regulation of Replication Origin Activity
The Initiation of DNA Replication in Eukaryotes
·
01 Jan 2016
·
doi:10.1007/978-3-319-24696-3_6
2015
Discovery of an Unconventional Centromere in Budding Yeast Redefines Evolution of Point Centromeres
Current Biology
·
01 Aug 2015
·
doi:10.1016/j.cub.2015.06.023
This work is H1 recommended.
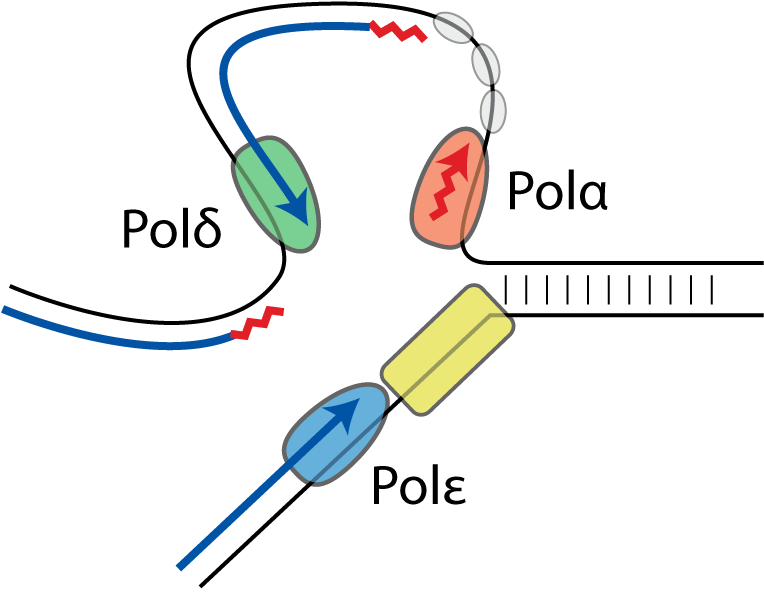
A global profile of replicative polymerase usage
Nature Structural & Molecular Biology
·
09 Feb 2015
·
doi:10.1038/nsmb.2962
This collaborative study reveals that polymerase epsilon is responsible for leading strand synthesis, with polymerase delta undertaking lagging strand synthesis. Our polymerase usage data directly maps replication origin location, quantifies origin efficiency, reveals that replication termination events are dispersed across large termination zones and indirectly estimates replication timing.
With associated News & Views by Sue Jinks-Robertson & Hannah L Klein.
With associated News & Views by Sue Jinks-Robertson & Hannah L Klein.
2014
Investigations in Yeast Functional Genomics and Molecular Biology
Apple Academic Press
·
24 Feb 2014
·
doi:10.1201/b16586
2013
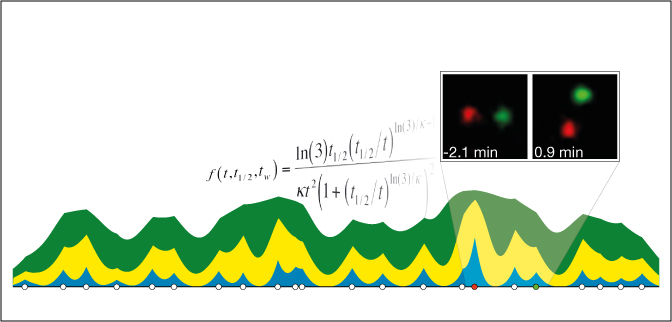
High-Resolution Replication Profiles Define the Stochastic Nature of Genome Replication Initiation and Termination
Cell Reports
·
01 Nov 2013
·
doi:10.1016/j.celrep.2013.10.014
In this study we used a combination of high-resolution, genomic-wide DNA replication data, mathematical modelling and single cell experiments to reveal the stochastic of replication initiation and termination. We find that the homogeneous picture implied by ensemble datasets masks biologically significant cell-to-cell variation in genome replication.
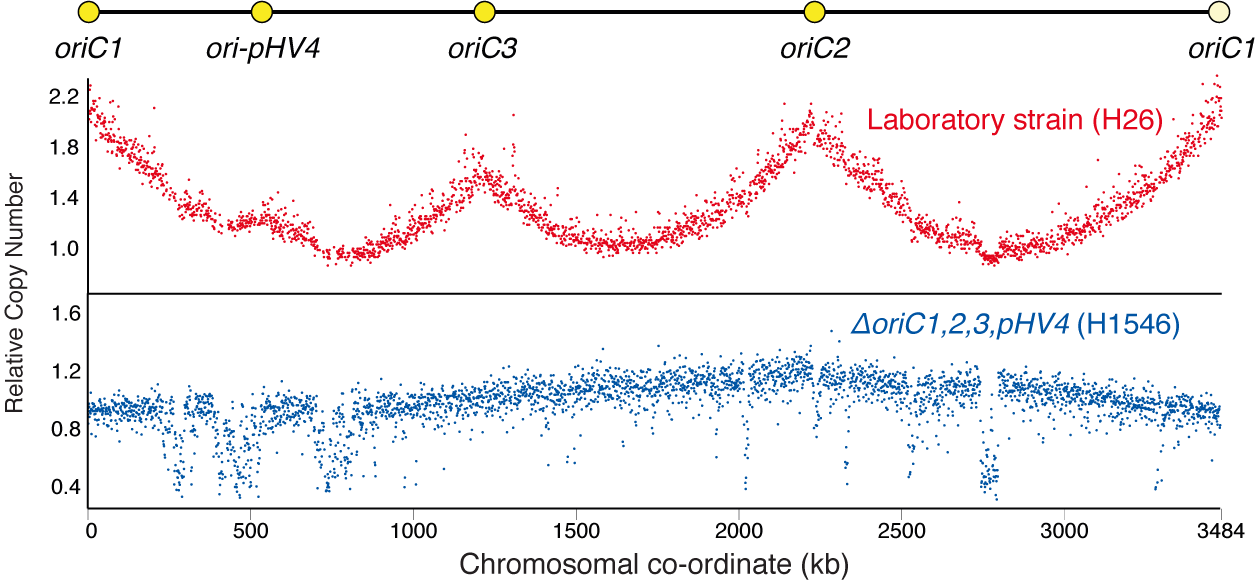
Accelerated growth in the absence of DNA replication origins
Nature
·
01 Nov 2013
·
doi:10.1038/nature12650
We discovered that the halophilic archaea, Haloferax volcanii, not only can survive in the absence of DNA replication origins, but also has accelerated growth.
This work is H1 recommended and featured in a research highlight in Nature Reviews Microbiology.
See associated article at The Conversation.
This work is H1 recommended and featured in a research highlight in Nature Reviews Microbiology.
See associated article at The Conversation.
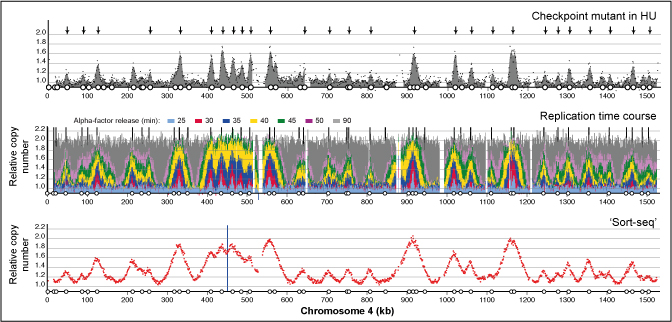
The dynamics of genome replication using deep sequencing
Nucleic Acids Research
·
01 Oct 2013
·
doi:10.1093/nar/gkt878
This paper describes and rigorously validates a range of next generation sequencing approaches to reveal at unprecedented resolution the temporal order of genome replication. My research group and others are applying these sequencing approaches to a range of questions in a variety of organisms.
Stochastic association of neighboring replicons creates replication factories in budding yeast
Journal of Cell Biology
·
23 Sep 2013
·
doi:10.1083/jcb.201306143
A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
PLoS Genetics
·
12 Sep 2013
·
doi:10.1371/journal.pgen.1003798
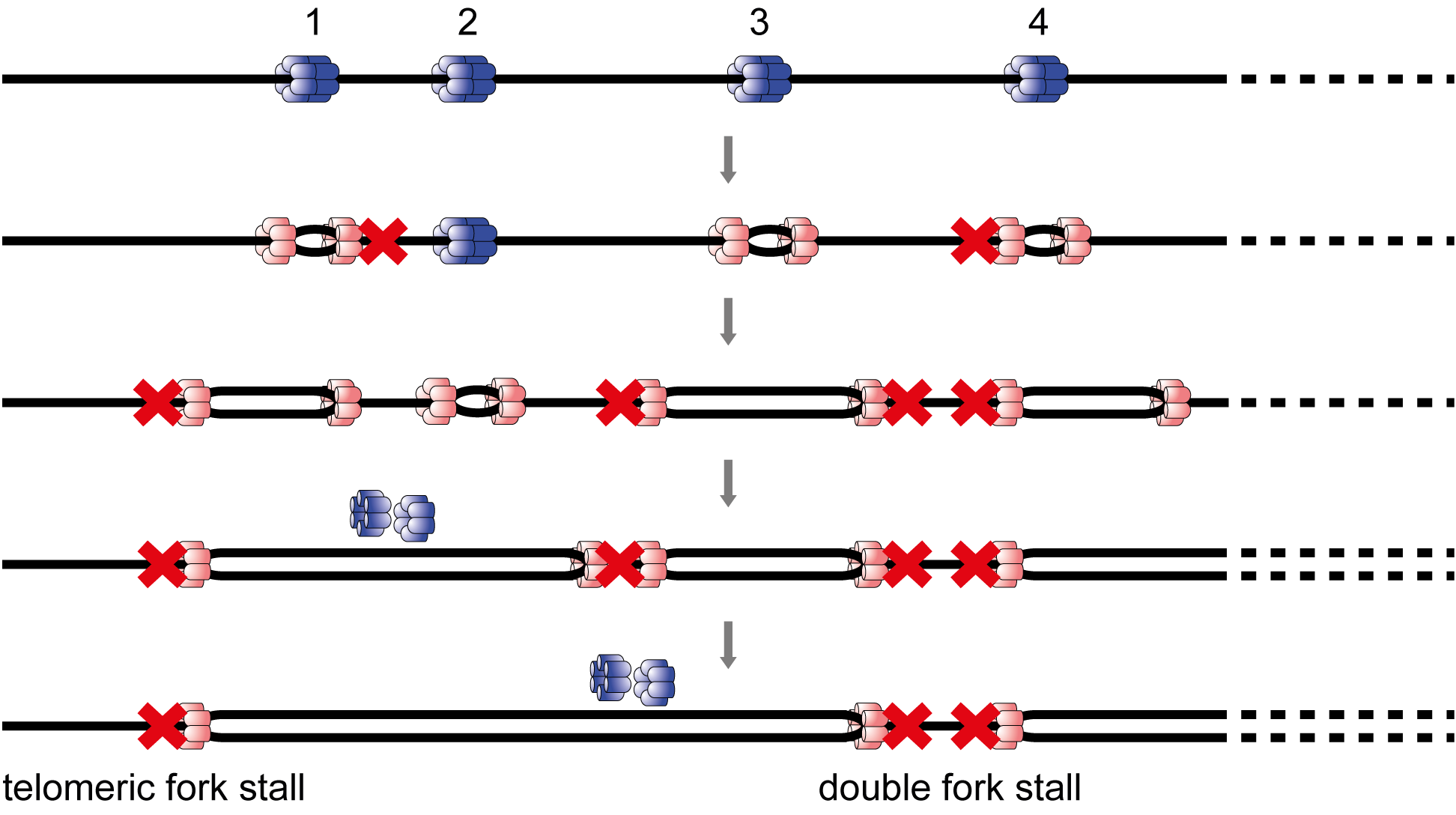
Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts
Nucleic Acids Research
·
19 Aug 2013
·
doi:10.1093/nar/gkt728
Avoiding chromosome pathology when replication forks collide
Nature
·
28 Jul 2013
·
doi:10.1038/nature12312

Kinetochores Coordinate Pericentromeric Cohesion and Early DNA Replication by Cdc7-Dbf4 Kinase Recruitment
Molecular Cell
·
01 Jun 2013
·
doi:10.1016/j.molcel.2013.05.011
We discovered that kinetochore recruitment of the Dbf4-dependent kinase ensures early centromere replication. Mutations that delay centromere replication give rise to elevated chromosome loss, revealing the first physiological role for temporal regulation of genome replication.
This work is H1 recommended.
This work is H1 recommended.
High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome
BMC Genomics
·
01 Jan 2013
·
doi:10.1186/1471-2164-14-69
2012
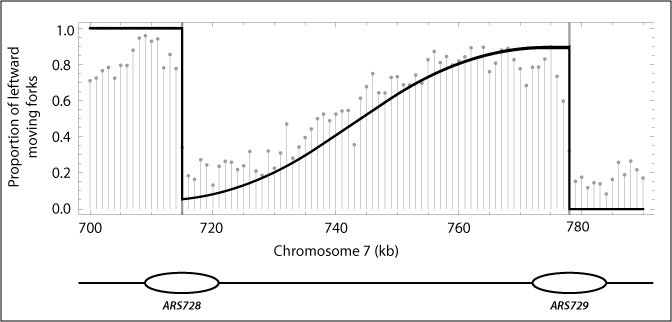
Mathematical modeling of genome replication
Physical Review E
·
17 Sep 2012
·
doi:10.1103/PhysRevE.86.031916

Conservation of replication timing reveals global and local regulation of replication origin activity
Genome Research
·
05 Jul 2012
·
doi:10.1101/gr.139477.112
We found that the temporal order of genome replication is conserved between species. The discovery of a minority of DNA replication origins that have evolved different activities allowed us to demonstrate that origin activity is locally (cis) regulated.
A Putative Homologue of CDC20/CDH1 in the Malaria Parasite Is Essential for Male Gamete Development
PLoS Pathogens
·
23 Feb 2012
·
doi:10.1371/journal.ppat.1002554
2011

OriDB, the DNA replication origin database updated and extended
Nucleic Acids Research
·
24 Nov 2011
·
doi:10.1093/nar/gkr1091
From sequence to function: Insights from natural variation in budding yeasts
Biochimica et Biophysica Acta (BBA) - General Subjects
·
01 Oct 2011
·
doi:10.1016/j.bbagen.2011.02.004
Dynamics of DNA Replication in Yeast
Physical Review Letters
·
04 Aug 2011
·
doi:10.1103/PhysRevLett.107.068103
This work is H1 recommended.
Comparative Functional Genomics of the Fission Yeasts
Science
·
20 May 2011
·
doi:10.1126/science.1203357
This work is H1 recommended.
2010
Mathematical modelling of whole chromosome replication
Nucleic Acids Research
·
10 May 2010
·
doi:10.1093/nar/gkq343
We present a mathematical model for whole chromosome replication, conceptualising how key molecular properties of DNA replication are related and demonstrating the wealth of information available in quantitative genomic datasets.
This work is H1 recommended.
This work is H1 recommended.
2009
The Origin Recognition Complex Interacts with a Subset of Metabolic Genes Tightly Linked to Origins of Replication
PLoS Genetics
·
04 Dec 2009
·
doi:10.1371/journal.pgen.1000755
Detection of Replication Origins Using Comparative Genomics and Recombinational ARS Assay
Methods in Molecular Biology
·
01 Jan 2009
·
doi:10.1007/978-1-60327-815-7_16
2008
Analysis of Chromosome III Replicators Reveals an Unusual Structure for the ARS318 Silencer Origin and a Conserved WTW Sequence within the Origin Recognition Complex Binding Site
Molecular and Cellular Biology
·
01 Aug 2008
·
doi:10.1128/MCB.00206-08
This work is H1 recommended.
2006

OriDB: a DNA replication origin database
Nucleic Acids Research
·
07 Nov 2006
·
doi:10.1093/nar/gkl758
This paper presents the DNA replication origin database, OriDB, a catalogue of confirmed and predicted DNA replication origin sites. This database is an example of our commitment not only to sharing data, but also to making it highly accessible to the research community – see links on the resources page.
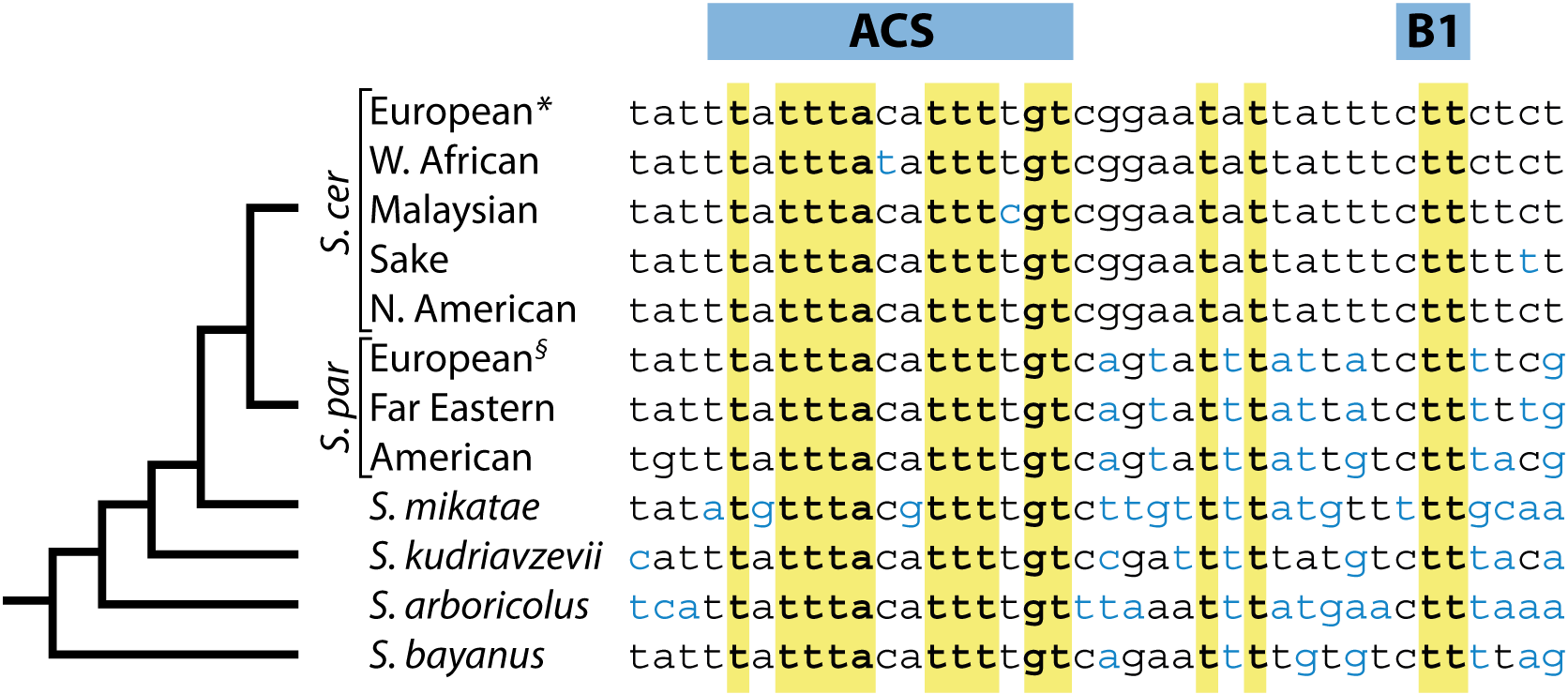
Genome-wide identification of replication origins in yeast by comparative genomics
Genes & Development
·
15 Jul 2006
·
doi:10.1101/gad.385306
This was the first paper to use comparative genomics to investigate DNA replication. We discovered that the functional sequence elements at DNA replication origins are evolutionarily conserved, leaving a phylogenetic footprint that allowed them to be pin- pointed genome-wide.
This work is H1 recommended.
This work is H1 recommended.
2005
The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription
Nucleic Acids Research
·
28 Apr 2005
·
doi:10.1093/nar/gki539
2004
The Cyclin A1-CDK2 Complex Regulates DNA Double-Strand Break Repair
Molecular and Cellular Biology
·
01 Oct 2004
·
doi:10.1128/MCB.24.20.8917-8928.2004
Cyclin A1 protein shows haplo-insufficiency for normal fertility in male mice
Reproduction
·
01 Apr 2004
·
doi:10.1530/rep.1.00131
2002
Whole-genome analysis of animal A- and B-type cyclins
Genome Biology
·
15 Nov 2002
·
doi:10.1186/gb-2002-3-12-research0070
Ku complex controls the replication time of DNA in telomere regions
Genes & Development
·
01 Oct 2002
·
doi:10.1101/gad.231602

Chromosome replication: from ORC to fork
Genome Biology
·
[no date info]
·
doi:10.1186/gb-2001-2-12-reports4030