Research projects

Initiation of DNA replication
Mapping DNA replication origins
The sites of DNA replication initiation, called replication origins, are spatially and temporally regulated. We have mapped the sites and characteristics of replication origins across the genome of several model organisms.
DNA replication dynamics
From initiation, to fork direction and finally termination
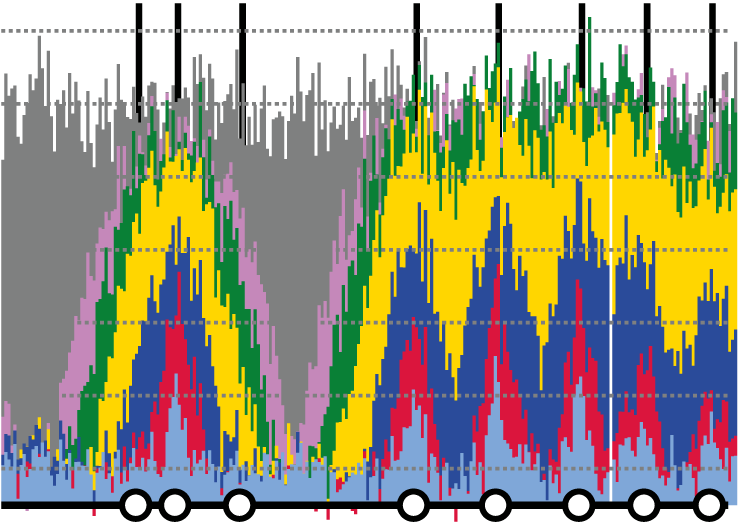
Genomes are replicated with characteristic temporal dynamics: some regions tend to replicate early in S phase whereas others tend to replicate later. We have measured replication dyanamics, discovered regulatory mechanisms and determined the consequences of perturbed replication dynamics.
Replication fork progression
Difficult to replicate sequences
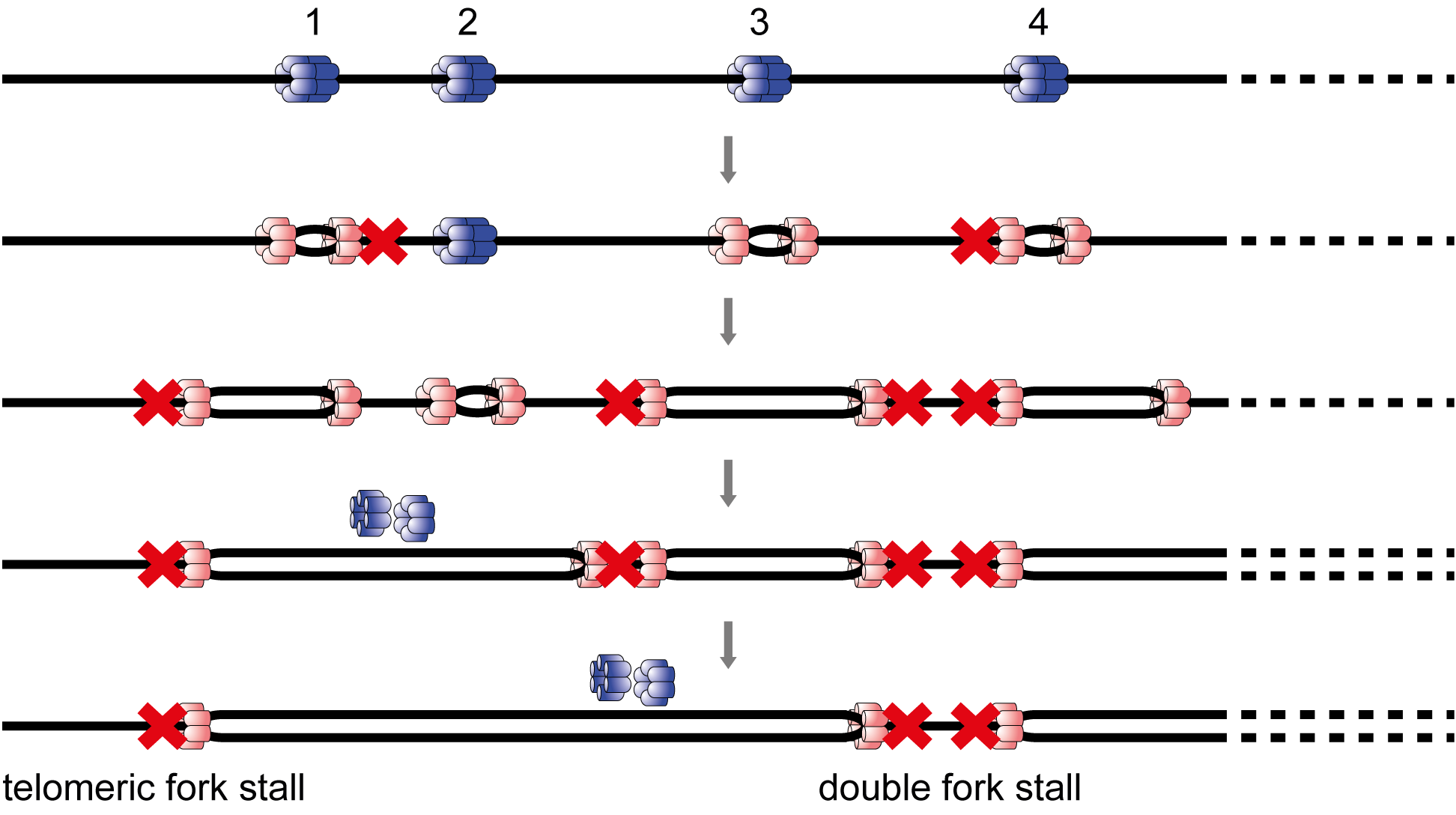
Genome replication is remarkably accurate and processive. However, some regions of the genome are more difficult to replicate. This can result in replication fork pausing and the potential for genome instability, for example from fork collapse or incomplete genome replication.
Synthetic genomes
Designing stable synthetic chromosomes
The Sc2.0 project has successfully synthesised all 16 native chromosomes in Saccharomyces cerevisiae. As part of this international collaboration, we have contributed to the design of synthetic chromosomes to ensure that they are faithfully replicated in cells.

Genome sequencing
Completing genomes
The power of genomic technologies are built upon high quality complete reference genome sequences. We have contributed to a series of high quality telomere-to-telomere genome sequences.

Genomic technologies
From populations to single molecules
We have developed a series of innovative genomic technolgies to determine DNA replication dynamics. Both population-level and single molecule approaches have allowed discovery of sites of replication initiation and subsequent replication dynamics.